Transmission dynamics of COVID-19
The pandemic of the Coronavirus disease 2019 (COVID-19), caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has been and will continue to be a challenge. Better understanding the dynamics of its spread can help meeting this challenge. A way to quantify the dynamics of an infectious disease is to determine specific epidemiological parameters. These include the reproductive number R, which stands for the number of secondary infections caused by one patient. For COVID-19, which can be asymptomatic, the number of missed cases is of special interest. Using phylodynamic methods, these parameters are quantified not only from incidence data, but mainly from the genetic sequence of isolated viruses.
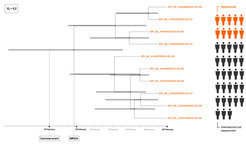
With this project, we quantify epidemiological parameters of the COVID-19 pandemic using birth-death models. A birth-death process describes the generation of a transmission tree forward in time through consecutive birth, death or sampling events. Under these models, we can determine three important epidemiological parameters: the effective reproductive number R_e, the recovery rate δ and the sampling proportion ψ, which gives a direct quantification of the number of missed cases. To learn more about the dynamics of COVID-19 we apply these methods to publicly available sequencing data from local outbreaks, which are epidemiologically linked, as well as region-wide data containing samples from many different transmission chains.
